
CA, which is clonally related to its adjacent glandular
adenocarcinoma
[11]. IDC/CA+ prostate cancers are strongly
associated with two other known adverse molecular
indices: genomic instability (chromothripsis and CNA)
and
SChLAP1
expression. In particular,
SChLAP1
was
singularly expressed at
>
3-fold in IDC/CA+ than in IDC/
CA– tumors. These molecular indices have an additive effect
on the lethality of these subpathologies. Combinatorial
indices of IDC/CA with either PGA (as a proxy for genomic
instability) or
SChLAP1
expression stratified patients for
recurrence more accurately than any parameter alone.
Taken together, the co-occurrence of multiple unfavorable
characteristics lends itself to a concept that closely
resembles
nimbosus
(‘‘gathering of stormy clouds’’; Latin),
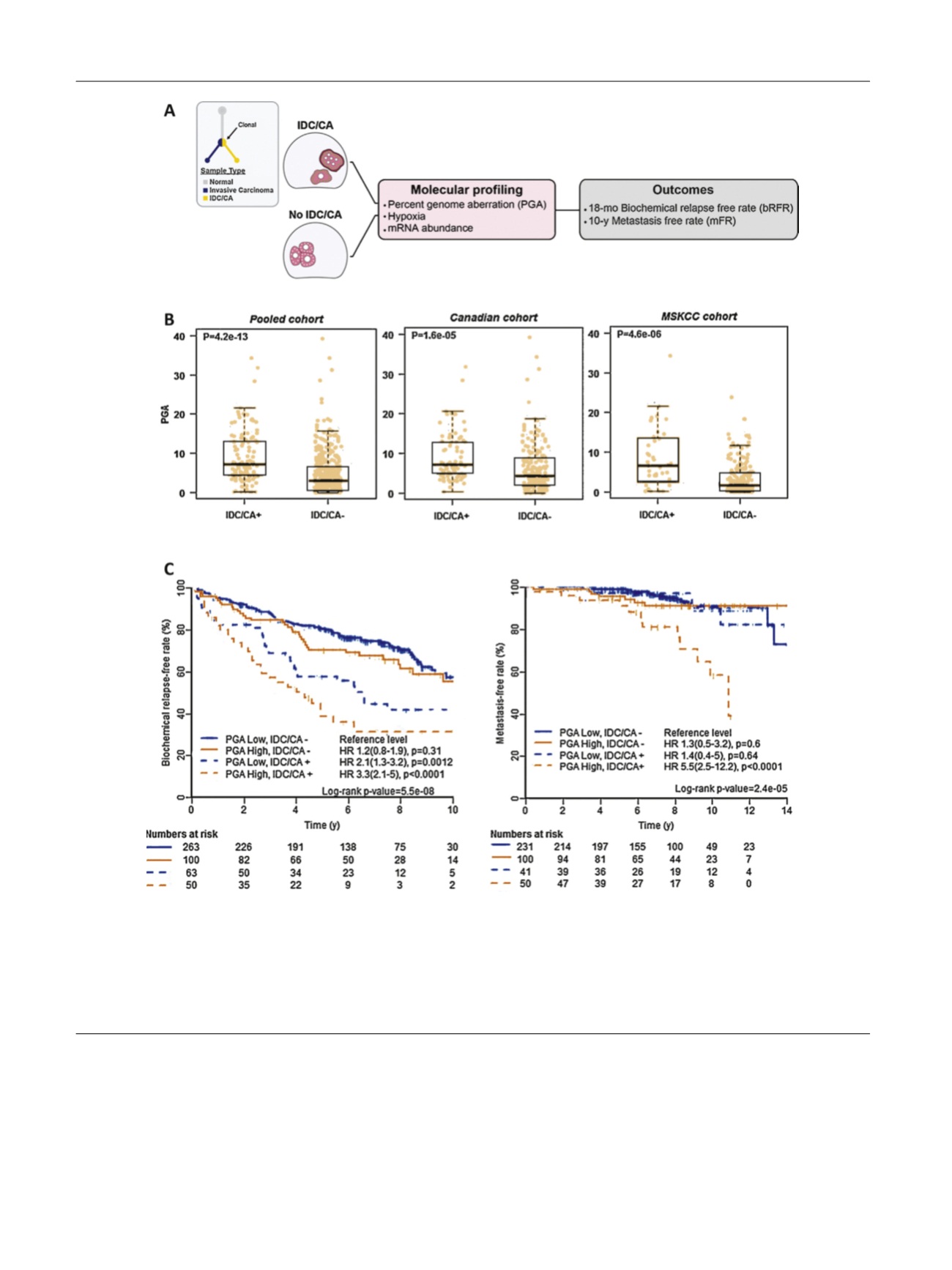
[(Fig._2)TD$FIG]
Fig. 2 – Outline of study and interactions between subpathologies and percentage of genome alteration (PGA). (A) To test for ‘‘aggression’’ field defect
in clonal intraductal carcinoma (IDC), cribriform architecture (CA), and adjacent glandular adenocarcinoma, we characterized the association between
IDC/CA+ prostate tumors and known adverse indices, and their association with biochemical and metastatic relapses. (B) Box and whisker (first/third
quartiles
W
interquartile range) plots of PGA in IDC/CA+ and IDC/CA– tumors for the pooled (left), Canadian (middle), and MSKCC (right) cohorts. (C)
Biochemical relapse- and metastasis-free rates based on combinatorial pathological and genomic stratification in 476 men with NCCN-defined low- to
high-risk prostate cancer. CHUdeQ-UL cohort (
n
= 54, with pathological and genomic data) was excluded from analyses of metastasis-free rate, as no
metastasis event was recorded in this subset of cases. CHUdeQ-UL = CHU de Que´bec-Universite´ Laval; HR = hazards ratio; MSKCC = Memorial Sloan
Kettering Cancer Center; NCCN = National Comprehensive Cancer Network.
E U R O P E A N U R O L O G Y 7 2 ( 2 0 1 7 ) 6 6 5 – 6 7 4
670
















